BoolODE¶
Overview¶
The BoolODE package converts a Boolean model to an ODE model and carries out stochastic simulations. It reads a model definition file, and outputs the simulated expression data for a given model.
python src/BoolODE.py --path=data/test_vars.txt --max-time=5 --num-cells 10
Tutorial¶
Defining an input model¶
Consider a toy gene regulatory network with a series of mutual inhibition motifs as illustrated below:
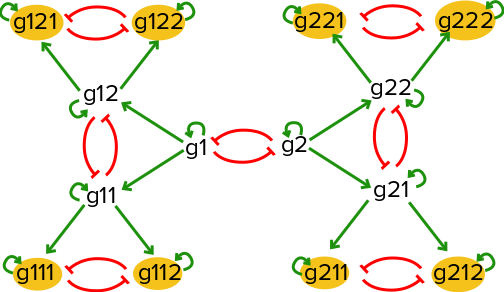
If there is an equal probabiliy of taking either the left hand branch or the right hand branch starting at (g1=high,g2=high), this network will result in 8 distinct steady states, with one of the eight extreme genes being active in each steady state.
BoolODE can be used to generate gene expression data of a sample of cells undergoing a developmental process controlled by a gene regulatory network with a wiring diagram as shown above.
The above network diagram can be represented as a Boolean model as follows:
Gene Rule
g1 ( g1 ) and not ( g2 )
g2 ( g2 ) and not ( g1 )
g11 ( g1 or g11 ) and not ( g12 )
g12 ( g1 or g12 ) and not ( g11 )
g21 ( g2 or g21 ) and not ( g22 )
g22 ( g2 or g22 ) and not ( g21 )
g111 ( g11 or g111 ) and not ( g112 )
g112 ( g11 or g112 ) and not ( g111 )
g121 ( g12 or g121 ) and not ( g122 )
g122 ( g12 or g122 ) and not ( g121 )
g211 ( g21 or g211 ) and not ( g212 )
g212 ( g21 or g212 ) and not ( g211 )
g221 ( g22 or g221 ) and not ( g222 )
g222 ( g22 or g222 ) and not ( g221 )
In order to test BoolODE, copy the rules above into a file, say multistate.txt,
in the /data folder in the BoolODE folder.
Note
BoolODE expects a tab separated rule file! Make sure the
input file has tabs between the Gene and Rule column.
Notice that the g1<->g2 interaction is central to the behavior of
the model. Let us choose an initial condition such that these genes
have a medium/high value. Biologically, this can be interpreted as the
initial undifferentiated cells have genes g1 and g2 ‘ON’, and
all other genes ‘OFF’. We can specify this initial state by creating a file with the
following lines:
Genes Values
['g1','g2'] [1,1]
The first column takes a list of genes formatted like a python list of strings. The second column takes a list of values to assign to the list of genes, again formatted as a python list of values. Any gene not listed in this file will be assigned an initial value of 0 by default.
Let us save the initial condition specification in
/data/multistate_ics.txt.
Note
The initial condition file should also be tab separated. Also, pay attention to the capitalization of the column names.
Tip
We recommend starting off with initial condition values between 0 and 2, since larger values might cause numerical instability.
Running BoolODE¶
A few more decisions before we get our simulated dataset of gene expression values:
- How long do you want to simulate this model? Let’s choose 8 time units
- How many cells do you want to simulate? 2000 seems like a good number.
- Where would you like to store the simulation output? Say,
./test/. BoolODE will create any folders that don’t already exist. - Do you want to speed up the simulations by running simulations in parallel? Yes!
One last option that we will add is --sample-cells, which will instruct BoolODE
to sample one cell from each simulated trajectory.
This is all we need to call BoolODE on this model! The following directive puts together all the information collected so far
python src/BoolODE.py --path data/multistate.txt \ # path to the Boolean rules file
--ics data/multistate_ics.txt \ # path to initial condition specification
--max-time 8 \ # maximum time of simulation
--num-cells 2000 \ # number of cells/simulations to perform
--outPrefix test/ \ # relative path to destination
--do-parallel \ # do parallel simulations
--sample-cells # sample one cell from each simulation
For a full list of available options, see Command Line Options
Working with BoolODE output¶
The simulations will take about 5 minutes to complete. At the end of a successful run, the output directory should like this:
test/ # User specified destination
|-- parameters.txt # Parameter names and values generated for input model
|-- ExpressionData.csv # Gene expression dataset
|-- PseudoTime.csv # Simulation time of each sample time point/cell
|-- refNetwork.csv # Boolean network represented as an edge list, the ground truth network
`-- simulations/
|-- E1.csv
|-- E2.csv
...
Where E1.csv, E2.csv, ... are individual simulations. Each column in these
files, the cell IDs, has the form E<simulation number>_<timepoint>.
Note
By default BoolODE will store the entire simulated time
course for every simulation. Specifying the --sample-cells
option will result in BoolODE sampling cells and creating an
ExpressionData.csv file with genes as rows, and cells as columns.
If the option is not specified, you can write a custom script to
sample cells from each simulation
In order to visualize the entire dataset, we can carry out dimensionality reduction using t-SNE. A script like the one below is a good starting point for this.
import pandas as pd
from sklearn.manifold import TSNE
import matplotlib
import matplotlib.pyplot as plt
def vis(df, p):
tsne = TSNE(n_components=2,perplexity=p).fit_transform(df.T.values)
tdf = pd.DataFrame(tsne, columns=['t-SNE 1', 't-SNE 2'],index=df.columns)
tdf.to_csv('test/tsne.csv')
fig, ax = plt.subplots(1,1,figsize=(5,5))
ax.scatter(tsne[:,0], tsne[:,1], c = [float(col.split('_')[1]) for col in df.columns])
ax.axis('off')
plt.tight_layout()
plt.savefig('test/tree.png')
df=pd.read_csv('test/ExpressionData.csv', index_col=0)
vis(df, 400)
Here, the time point of each cell is inferred from the cell ID, and this information is used to color each cell in the scatter plot. Darker colors imply early time points in the simulation. The output should look like the following
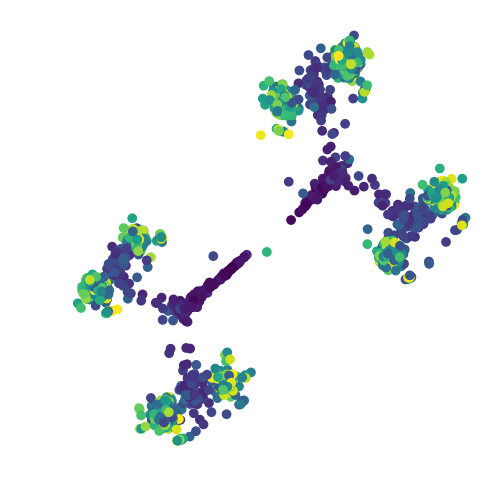
t-SNE visualization of BoolODE output
Notice the eight steady state clusters! This dataset can now be processed further using the tools described in Generating inputs for BEELINE to produce input datasets for the BEELINE pipeline.
Generating inputs for BEELINE¶
BoolODE provides additional scripts, located in scripts/ to
process simulation output to be used by BEELINE.
Compute Pseudotime using Slingshot¶
Note
runSlingshot.py requires the SlingShot docker file! Please make sure docker has been setup and the container built.
Slingshot computes pseudotime trajectories for a given dataset by
first carrying out dimensionality reduction, then carrying out
k-means clustering on the low dimensional embedding in order to
compute trajectories. The number of clusters expected depend on the
features of the dataset. For instance, a dataset with two steady state
clusters should be specified with --nClusters 3, specifying an
additional initial state cluster.
runSlingshot.pytakes the following command line arguments-h, --help show this help message and exit --outPrefix=OUTPREFIX Prefix for output files. -e EXPR, --expr=EXPR Path to expression data file -p PSEUDO, --pseudo=PSEUDO Path to ‘pseudotime’ file generated by BoolODE -c NCLUSTERS, --nClusters=NCLUSTERS Number of expected clusters in the dataset. --noEnd Do not force SlingShot to have an end state. -r PERPLEXITY, --perplexity=PERPLEXITY Perplexity for tSNE.
Note
runslingshot.py requires a ‘pseudotime’ file passed
using the --pseudo option. This file should contain the
actual simulation time from BoolODE, which can then be used
to compare the quality of the inferred trajectory with the
actual simulation time values. This file is NOT required by
Slingshot itself.
Generate dropouts from expression data¶
In order to mimic real single-cell expression datasets, BoolODE
includes genDropouts.py which implements dropouts as described in
the paper, by dropping expression values below DROP_CUTOFF using a
probability of DROP_PROB. This script samples NCELLS from the
columns in the expression dataset EXPR, and will throw an error if
the number is greater than the number of columns.
genDropouts.pytakes the following command line arguments-h, --help show this help message and exit --outPrefix=OUTPREFIX Prefix for output files. -e EXPR, --expr=EXPR Path to expression data file -p PSEUDO, --pseudo=PSEUDO Path to pseudotime file -r REFNET, --refNet=REFNET Path to reference network file -n NCELLS, --nCells=NCELLS Number of cells to sample. -d, --dropout Carry out dropout analysis? [Optional] --drop-cutoff=DROP_CUTOFF Specify percentile cutoff on gene expression --drop-prob=DROP_PROB Specify the probability of dropping a gene below quantile q. Ensure 0 < DROP_PROB < 1. -i SAMPLENUM, --samplenum=SAMPLENUM Sample Number
Attention
Ensure the --dropout option is passed. If not, genDropouts
will still randomly sample cells but not dropout any values.
Command Line Options¶
-h, --help show this help message and exit --max-time=MAX_TIME Total time of simulation. (Default = 20) --num-cells=NUM_CELLS Number of cells sample. (Default = 100) --sample-cells Sample a single cell from each trajectory? By default will store full trajectory of each simulation (Default = False) --add-dummy Add dummy genes -n, --normalize-trajectory Min-Max normalize genes across all experiments -i, --identical-pars Set single value to similar parameters NOTE: Consider setting this if you are using –sample-pars. -s, --sample-pars Sample rate parameters around the hardcoded means , using 10% stand. dev. --std=STD If sampling parameters, specify standard deviation. (Default 0.1) --write-protein Write both protein and RNA values to file. Useful for debugging. -b, --burn-in Treats the first 25% of the time course as burnin , samples from the rest. --outPrefix=OUTPREFIX Prefix for output files. --path=PATH Path to boolean model file --inputs=INPUTS Path to input parameter files. This is different from specifying a parameter set! --pset=PSET Path to pre-generated parameter set. --ics=ICS Path to list of initial conditions --strengths=STRENGTHS Path to list of interaction strengths --species-type=SPECIES_TYPE Path to list of molecular species type file.Useful to specify proteins/genes -c NCLUSTERS, --nClusters=NCLUSTERS Number of expected clusters in the dataset. (Default =1) --max-parents=MAX_PARENTS Number of parents to add to dummy genes. (Default = 1) --do-parallel Run simulations in parallel. Recommended for > 50 simulations